Staining Dye, Loading Dye, and Gel Stains
FluoroStain™ DNA Fluorescent Staining Dye (DS1000)
1. Will FluoroStain™ DNA Fluorescent Staining Dye (DS1000) affect the DNA samples in subsequent experiments?
Using FluoroStain™ DNA Fluorescent Staining Dye does not affect subsequent operations. This is because the fluorescent dye can easily be removed by regular alcohol precipitation or a gel elution kit.
2. Will the FluoroStain™ DNA Fluorescent Staining Dye (DS1000) affect cloning efficiency?
Using fluorescent dye coupled with blue light, the cloning efficiency is increased by about 100 times compared to using EtBr and UV light.
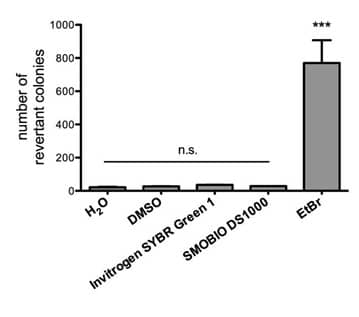
3. Are there relevant documents that support the safety of FluoroStain™ DNA Fluorescent Staining Dye (DS1000)?
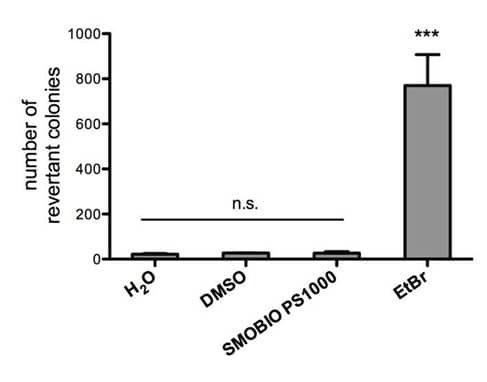
FluoroStain™ DNA Fluorescent Staining Dye (DS1000) is proofed for safety (non-mutagenicity) using the Ames test (Figures below). However, it must be noted that since solvents may penetrate the skin, it is recommended that users wear gloves when using the fluorescent dyes.
4. Is it possible to use FluoroStain™ DNA Fluorescent Staining Dye (DS1000) instead of ethidium bromide (EtBr) in staining of DGGE gels which is composed of poly acrylamide?
In the case of DGGE staining with FluoroStain™ DNA Fluorescent Staining Dye (DS1000), we can give some suggestions as stated below:
A. Like ethidium bromide, DS1000 could be used for staining DNA in polyacrylamide gels.
B. However, it is noted that the DS1000 is designed to stain dsDNA and thus is not appropriate for staining in DGGE due to denaturing of dsDNA.
C. For detection of ssDNA or RNA in DGGE, we strongly recommend the use of FluoroVue™ Nucleic Acid Gel Stain (NS1000) which can be used to stain both double-strands and single-strand nucleic acids.
D. Furthermore, it is recommended to stain the gel after electrophoresis (post-staining) instead of pre-staining or in-gel staining, since the fluorescent dye might be affected during acrylamide polymerization.
E. The optimal excitation wavelength of DS1000 or NS1000 is around 480~490 nm (blue light), and therefore the illuminator with blue light is preferred. Of course, UV could be used also, but the signals detected may be weaker.
5. In using FluoroStain™ DNA Fluorescent Staining Dye (DS1000), which method is recommended as the gel soaking and in-gel staining methods (pre-casted gel with fluorescent dye) are considered?
FluoroStain™ DNA Fluorescent Staining Dye (DS1000) should be used by gel soaking (stain the gel after electrophoresis). For in-gel stain, FluoroVue™ Nucleic Acid Gel Stain (NS1000) is suggested
6. After post staining using the FluoroStain™ DNA Fluorescent Staining Dye (DS1000), why arethe DNA bands sometimes blurred and the fluorescent signal is not enough?
There are three possible reasons:
A. The fluorescent dye is very sensitive, so a small amount of smear due to DNA quality may blur the band.
B. The agarose is impure, or incompletely dissolved. Tailing effect may also cause the fluorescent signal to weaken.
C. There may not be enough dye or the staining time is insufficient. After each staining, gels will absorb a large amount of fluorescent dye, thus causing insufficient staining for the next time if the same staining bath is repeatedly used. Furthermore, the time required for staining can be elongated based on the percentage and the thickness of the agarose gel; harder and thicker gels require a longer staining period.
7. Do you recommend using DNA staining dye diluted in loading buffer?
We do not recommend this because the wrong concentration after dilution may cause inconsistent results.
8. Does the quality of agarose gel matter when using FluoroStain™ DNA fluorescent staining dye (DS1000)?
Yes, insufficient quality of agarose gel may lead to the limited performance of the dye.
9. What is the generally recommended dilution of FluoroStain™ DNA fluorescent staining dye (DS1000)?
A dilution of 10,000 times. For example, 10 μL staining dye is added to 100 mL of water, TE, TAE, or TBE. Next soak the agar gel in the solution. Soaking time depends on the percentage and thickness of agarose gel.
10. Does using FluoroStain™ DNA Fluorescent Staining Dye (DS1000) for staining the DNA cause bias in DNA molecular weight determination?
No, when the gel is stained with FluoroStain™ DNA Fluorescent Staining Dye (DS1000) after electrophoresis, the DNA molecular weight interpretation is accurate.
11. How many ways can the FluoroStain™ DNA Fluorescent Staining Dye (DS1000) be used?
For using the FluoroStain™ DNA Fluorescent Staining Dye (DS1000), it is recommended to be used with apost staining method. If DS1000 is used with in-gel staining or staining during electrophoresis, the DNA band may not be clear enough and the DNA migration may be changed.
12. Why does the gel stained with FluoroStain™ DNA Fluorescent Staining Dye (DS1000) show minimal background which is barely seen as compared to DNA signals that are strongly fluorescent after staining?
FluoroStain™ DNA Fluorescent Staining Dye (DS1000) manifests fluorescent signal when it properly binds with double stranded DNA.
13. Can FluoroStain™ DNA Fluorescent Staining Dye (DS1000) be used to observe RNA?
FluoroStain™ DNA Fluorescent Staining Dye (DS1000) is designed specifically for double stranded DNA staining and is not for RNA use. If there is the need to stain RNA, it is recommended to use FluoroVue™ nucleic acid stain (NS1000) to stain RNA.
14. During the dilution of FluoroStain™ DNA Fluorescent Staining Dye (DS1000), it is occasionally seen that there are a few red flocculent unable to be completely dissolved in the buffer. Is there any method to improve this?
To avoid the solid precipitation of FluoroStain™ DNA Fluorescent Staining Dye (DS1000) during -20℃ (4℃) storage, we recommend opening the storage tubes only when the DS1000 solution is complete melted.
If solid precipitation of DS1000 was observed, warm the tube at 37℃ for 10 min and then vortex to dissolve the solid particles. If the solid precipitation of DS1000 was still unable to be completely dissolved, spin down the precipitate and use only supernatant to dilute in TAE/TBE/TE buffer. A few solid precipitations of DS1000 will not cause the decline of sensitivity.
15. FluoroStain™ DNA Fluorescent Staining Dye (DS1000) Retention period: Mentioned -20℃ and over 2 years. Is it possible to have a retention period of 30 months or 36 months? Have you tested this period?
In our tests, the retention period of FluoroStain™ DNA Fluorescent Staining Dye (DS1000) could extend beyond 2 years if it is maintained at the recommended temperature of -20℃. However, we must recommend that the retention period should still be 2 years. Storage conditions should be in a dark room and the storage temperature kept at -20℃. High temperature and light will result in fluorescent dye attenuation. We have tested our DS1000 at -20℃ for 30 months and it did perform with similar efficiency. Again, we do not recommend a retention period of more than 24 months at -20℃.
16. How many cycles of freezing and thawing can the FluoroStain™ DNA Fluorescent Staining Dye (DS1000) handle? If 5 μL is used at once, will it have a usage of 100 times?
Our tests have shown that 100 freezing and thawing cycles will be appropriate for the FluoroStain™ DNA Fluorescent Staining Dye (DS1000). In DS1000's usage information, it is indicated that the fluorescent dye should be protected from light and kept at low temperatures. After using DS1000, it should immediately be kept between 4℃ to -20℃ as fluorescent dye decays at a faster rate at room temperature. Please remember that any un-thawed solution will cause the fluorescent dye concentration to be less after each use therefore reducing its sensitivity performance.
17. Is there any information on the ionic strength, pH stability, and optimum pH for FluoroStain™ DNA Fluorescent Staining Dye (DS1000)?
FluoroStain™ DNA Fluorescent Staining Dye (DS1000) stock is diluted in DMSO and does not have any special pH range. It is not advisable to use DS1000 in water. Therefore, it should be used with 1x TAE, TBE, and TE buffer at pH8.0 to gain the desired results. The optimum pH value for DS1000 is 7.5 to 8.0. Less than 7.5 and more than 8.0 may result in reduced sensitivity.
18. Can FluoroStain™ DNA Fluorescent Staining Dye (DS1000) be removed from DNA? How could this be done?
The staining dye can be removed from DNA with traditional ethanol precipitation, PCR clean up kits, or the gel extraction kits. The ethanol precipitation method can follow conventional molecular cloning or follow the listed protocol. Ethanol precipitation.
A. Measure the volume of the DNA sample.
B. Add 1/10 volume of 3M sodium acetate, pH 5.2,(final concentration of 0.3 M) - The pH value of 3M sodium acetate must be adjusted with acetate not with HCl.
C. Mix well.
D. Add 2 to 2.5 volumes of cold 100% ethanol (calculated after salt addition).
E. Mix well.
F. Place on ice or at -20 °C for >20 minutes.
G. Spin at maximum speed in a microfuge for 10-15 min.
H. Carefully decant supernatant.
I. Add 1 mL 70% ethanol. Rinse and spin briefly. Carefully decant supernatant.
J. Air dry or briefly vacuum dry pellet.
K. Re-suspend pellet in the appropriate volume of TE, Tris buffer or water.
19. Can FluoroStain™ DNA Fluorescent Staining Dye (DS1000) stained DNA be used for enzymatic reaction like ligation, enzyme cutting, PCR and so on? Do you have any recommended methods such as pretreatment?
After removing the fluorescent staining dye from DNA, the DNA can be used for ligation, restriction enzyme digestion or PCR reaction. It is imperative that the DNA is maintained in good quality for bioassay after removing the fluorescent staining dye. We recommend removing the fluorescent staining dye with the gel extraction kits or PCR clean up kits because these methods are convenient and high efficiency. The very low amount of fluorescent staining dye does not affect ligation, enzyme digestion or PCR. However, the threshold is related to the enzyme systems and it is on a case-by-case basis. For the best results, we recommend removing the staining dye from DNA before proceeding to the next step of the experiment.
20. How do we dispose of the staining solution? Is the disposal method for EtBr solution acceptable for this product?
We recommend that disposal be made in accordance with local laws. The freshly prepared FluoroStain™ DNA Fluorescent Staining Dye (DS1000) solution can be filtered through activated charcoal before disposal. The charcoal can then be disposed of by incineration. One gram of activated charcoal easily absorbs the dye from 10 liters of freshly prepared working solution.
21. What is the stability of the FluoroStain™ DNA Fluorescent Staining Dye (DS1000)? Is it stable in hot condition and does it use in-gel or post staining?
The FluoroStain™ DNA staining dye (DS1000) can be stored at -20℃ for at least 24 months. If it needs to be used frequently, it can be stored at room temperature or 4℃. The DS1000 is recommended to be used with post stain method. For in-gel staining the best choice would be to use FluoroVue™ Nucleic Acid Gel Stain (NS1000).
FluoroVue™ Nucleic Acid Gel Stain (NS1000)
1. Will FluoroVue™ Nucleic Acid Gel Stain (NS1000) affect the DNA/RNA samples in subsequent experiments?
Using FluoroVue™ Nucleic Acid Gel Stain does not affect subsequent operations. This is because the fluorescent dye can easily be removed with regular alcohol precipitation or gel elution kits.
2. Will the FluoroVue™ Nucleic Acid Gel Stain (NS1000) affect cloning efficiency?
Using the fluorescent dye coupled with blue light, the cloning efficiency is increased by about 100 times compared to using EtBr and UV light.
3. Are there relevant document that support the safety of FluoroVue™ Nucleic Acid Gel Stain (NS1000)?
FluoroVue™ Nucleic Acid Gel Stain (NS1000) is reviewed for safety, including non-mutagenicity and non-cytotoxicity (documents below). However, it must be noted that since solvents may penetrate the skin, it is recommended that users wear gloves when using the fluorescent dyes.
Safety report- Cytotoxicity test
4. Does the quality of agarose gel matter when using FluoroVue™ Nucleic Acid Gel Stain (NS1000)?
Yes, insufficient quality of agarose gel may lead to the limited performance of the dye.
5. How many ways can the FluoroVue™ Nucleic Acid Gel Stain (NS1000) be used?
For using the FluoroVue™ Nucleic Acid Gel Stain (NS1000), it is highly recommended to be used with in-gel staining method. NS1000 can also be used with post staining or staining during electrophoresis.
6. Why does the gel stained with FluoroVue™ Nucleic Acid Gel Stain (NS1000) show minimal background which is barely seen as compared with DNA/RNA signals that are strongly fluorescent after staining?
The FluoroVue™ Nucleic Acid Gel Stain (NS1000) exhibits fluorescent signals when it binds with nucleic acid (dsDNA, ssDNA, and RNA).
7. How many cycles of freezing and thawing can FluoroVue™ Nucleic Acid Gel Stain (NS1000) handle? If 5 μL is used at once, will it have a usage of 100 times?
Our tests have shown that 100 freezing and thawing cycles will be appropriate for the FluoroVue™ Nucleic Acid Gel Stain (NS1000). In NS1000's usage information, it is indicated that the fluorescent dye should be protected from light and kept at low temperatures. After using NS1000, it should immediately be kept between 4℃ to -20℃ as fluorescent dye decays at a faster rate at room temperature. Please remember that any un-thawed solution will cause the fluorescent dye concentration to be less after each use therefore decreasing its sensitivity performance.
8. Can FluoroVue™ Nucleic Acid Gel Stain (NS1000) be removed from DNA? If so, how can this be done?
The staining dye can be removed from DNA with traditional ethanol precipitation, PCR clean up kits, or the gel extraction kits. The ethanol precipitation method can follow conventional molecular cloning or follow the listed protocol. Ethanol precipitation.
A. Measure the volume of the DNA sample.
B. Add 1/10 volume of 3M sodium acetate, pH 5.2,(final concentration of 0.3 M) - The pH value of 3M sodium acetate must be adjusted with acetate not with HCl.
C. Mix well.
D. Add 2 to 2.5 volumes of cold 100% ethanol (calculated after salt addition).
E. Mix well.
F. Place on ice or at -20 °C for >20 minutes.
G. Spin at maximum speed in a microfuge for 10-15 min.
H. Carefully decant supernatant.
I. Add 1 mL 70% ethanol. Rinse and spin briefly. Carefully decant supernatant.
J. Air dry or briefly vacuum dry pellet.
K. Re-suspend pellet in the appropriate volume of TE, Tris buffer or water.
9. Can FluoroVue™ Nucleic Acid Gel Stain (NS1000) stained DNA be used for enzymatic reactions like ligation, enzyme digestion, PCR and so on? Do you have any recommended methods like pretreatment?
After removing the fluorescent staining dye from DNA, the DNA can be used for ligation, restriction enzyme digestion or PCR reaction. It is imperative that the DNA is maintained in good quality for bioassay after removing the stained dye. We recommend removing the fluorescent staining dye with the gel extraction kits or PCR clean up kits because these methods are convenient and high efficiency. The very low amount of fluorescent staining dye does not affect ligation, enzyme digestion or PCR. However, the threshold is related to the enzyme systems, and it is on a case-by-case basis. For the best results, we recommend removing the fluorescent staining dye from DNA before proceeding to the next step of the experiment.
10. How do we dispose of the staining solution? Is the disposal method for EtBr solution acceptable for this product?
We recommend that disposal be made in accordance with local laws. The freshly prepared DS1000 solution can be filtered through activated charcoal before disposal. The charcoal can then be disposed of by incineration. One gram of activated charcoal easily absorbs the dye from 10 liters of freshly prepared working solution.
11. When FluoroVue™ Nucleic Acid Gel Stain (NS1000) or EtBr is used for in-gel staining, why do some of the lowest bands become weaker in a longer agarose gel?
This is because EtBr or the dye component of NS1000 keep moving toward the cathode in a direction opposite to the DNA migration. Therefore, after electrophoresis the concentration of NS1000 or EtBr is very low near the anode end (the bottom) of the agarose gel, and some bands close to the bottom will be very weak in signal or even undetectable. We recommend reducing the electrophoresis time or using post staining methods to improve the intensity of small fragments of DNA.
12. How do we deal with certain precipitate in NS1000?
Occasionally, some precipitate will be observed in NS1000 due to the high concentration of dye ingredients.
If slight precipitate exists in NS1000, we suggest incubating the NS1000 at 37 ℃ for one hour until the precipitate is fully dissolved.
The sensitivity of NS1000 is maintained after the precipitate is dissolved.
FluoroDye™ DNA Fluorescent Loading Dye (DL5000)
1. Will FluoroDye™ DNA Fluorescent Loading Dye (DL5000) affect the DNA samples in subsequent experiments?
Using FluoroDye™ DNA Fluorescent Loading Dye (DL5000) does not affect subsequent operations. This is because the fluorescent dye can easily be removed by regular alcohol precipitation or gel elution kits.
2. Will FluoroDye™ DNA Fluorescent Loading Dye (DL5000) affect cloning efficiency?
Using the fluorescent dye coupled with blue light, the cloning efficiency is increased by about 100 times compared to using EtBr and UV light.
3. Can FluoroDye™ DNA Fluorescent Loading Dye (DL5000) be removed from DNA? How can we do it?
The staining dye can be removed from DNA with traditional ethanol precipitation, PCR clean up kits, or gel extraction kits. The ethanol precipitation method can follow conventional molecular cloning or follow the listed protocol. Ethanol precipitation.
A. Measure the volume of the DNA sample.
B. Add 1/10 volume of 3M sodium acetate, pH 5.2,(final concentration of 0.3 M) - The pH value of 3M sodium acetate must be adjusted with acetate not with HCl.
C. Mix well.
D. Add 2 to 2.5 volumes of cold 100% ethanol (calculated after salt addition).
E. Mix well.
F. Place on ice or at -20 °C for >20 minutes.
G. Spin at maximum speed in a microfuge for 10-15 min.
H. Carefully decant supernatant.
I. Add 1 mL 70% ethanol. Rinse and spin briefly. Carefully decant supernatant.
J. Air dry or briefly vacuum dry pellet.
K. Re-suspend pellet in the appropriate volume of TE, Tris buffer or water.
4. Can FluoroDye™ DNA Fluorescent Loading Dye (DL5000) stained DNA be used for enzymatic reactions such as ligation, enzyme digestion, PCR and so on? Do you have any recommended methods like pretreatment?
After removing the fluorescent staining dye from DNA, the DNA can be used for ligation, restriction enzyme digestion or PCR reaction. It is imperative that the DNA is maintained in good quality for bioassay after removing the stained dye. We recommend removing the fluorescent staining dye with the gel extraction kits or PCR clean up kits because these methods are convenient and high efficiency. The very low amount of fluorescent staining dye does not affect ligation, enzyme digestion or PCR. However, the threshold is related to the enzyme systems and it is on a case-by-case basis. For the best results, we recommend removing the fluorescent staining dye from DNA before proceeding to the next step of the experiment.
5. As the FluoroDye™ DNA Fluorescent Loading Dye (DL5000) is used, is the migration of DNA affected by the fluorescent dye?
Yes, it could be affected due to variation of DNA amount mixed with the fluorescent loading dye. The fluorescence composition in a fluorescent loading dye is fixed, so for different amounts of DNA different degrees of binding to fluorescent dyes are expected. Variety in DNA/fluorescence ratio might lead to variation in DNA migration; the higher the fluorescence ratio is, the slower the DNA fragments migrate. In general, a DNA amount of ≥31.25 ng with 1 μL DL5000 loading dye will be an acceptable range. If the DNA concentration is lower than 31.25 ng, the molecular weight determination may be slightly distorted. For analyzing DNA sample of unknown concentration, it is recommended to use the FluoroStain™ DNA staining dye with post-staining method or to use FluoroVue™ nucleic acid staining dye with in-gel staining method.
FluoroStain™ Protein Fluorescent Staining Dye (PS1000)
1. Can the SDS-PAGE gel stained with FluoroStain™ Protein Fluorescent Staining Dye (PS1000) be transferred to perform Western Blot? Will it affect the antibody binding when Western Blot is performed?
After using FluoroStain™ Protein Fluorescent Staining Dye (PS1000) to stain the gel, it is NOT recommended to perform Western Blot. The proteins in the SDS PAGE gel are fixed during staining with acidic solution, and thus preventing the transfer of proteins to NC or PVDF membrane.
2. When doing a follow up analysis using an SDS PAGE stained with FluoroStain™ Protein Fluorescent Staining Dye (PS1000), will there be any effect on mass analysis?
There will be no effect on mass analysis when following up with an SDS PAGE analysis using FluoroStain™ Protein Fluorescent Staining Dye (PS1000).
3. Is FluoroStain™ Protein Fluorescent Staining Dye (PS1000) able to stain a 2D gel?
The FluoroStain™ Protein Fluorescent Staining Dye (PS1000) can stain a 2D gel, which is quite sensitive. PS1000 can achieve detection levels parallel to silver staining. The sample gel can be used for further MASS analysis.
4. Are there relevant documents that support the safety (ex. non-mutagenicity) of FluoroStain™ Protein Fluorescent Staining Dye (PS1000)?
FluoroStain™ Protein Fluorescent Staining Dye (PS1000) is proofed for safety (non-mutagenicity) using the Ames test (Figures below). However, it must be noted that since solvent may penetrate the skin, it is recommended that users wear gloves when using the fluorescent dyes.